Abstract

Background: JAK2 V617F and exon 12 mutations are the characteristic driver mutations in polycythemia vera (PV), identified in more than 95% of patients. In addition, other genetic mutations have previously been described in JAK2-positive PV that appear to have prognostic significance (Tefferi et al., Blood 2016). The incidence of other driver mutations in unselected patients referred for elevated hemoglobin is less well studied. This study aims to characterize the genetic mutational landscape in a real-world population of patients referred for elevated hemoglobin using a targeted Next-Generation Sequencing (NGS)-based assay.
Methods: We reviewed all patients referred for elevated hemoglobin levels (>160 g/L in females or >165 g/L in males) between 2018 and 2020 to hematology clinics at London Health Sciences Centre in Southwestern Ontario, Canada who underwent testing for genetic variants using the NGS-based Oncomine Myeloid Research Assay (ThermoFisher Scientific, MA, USA). This assay targets 40 key genes with diagnostic and prognostic implications in several myeloid malignancies (17 full genes and 23 genes with clinically relevant "hotspot" regions) and a panel of 29 fusion driver genes (>600 fusion partners). Patient demographics, laboratory data and final diagnosis were extracted from the electronic medical record. For all patients with genetic mutations, clinical diagnosis was confirmed by three independent reviewers.
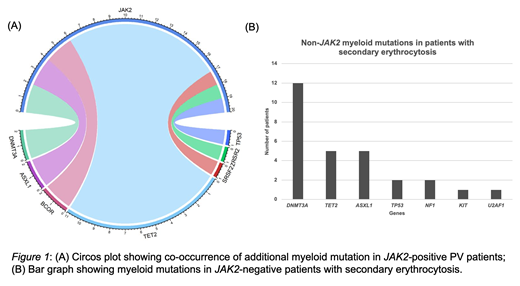
Results: A total of 529 patients underwent genetic testing for elevated hemoglobin levels: 389 (73.5%) were males (mean age 58; range 18-95) and 140 (26.5%) were female (mean age 60; range 24-85). JAK2 mutations were detected in 10.9% (58/529) of patients and a diagnosis of PV was confirmed. The majority of JAK2-mutated PV patients (n=57) were positive for JAK2 V617F, while one patient had an exon 12 mutation. Additional single myeloid mutations were detected in 34.5% (20/58) of JAK2-positive patients and involved the following genes: TET2 (11; 19%), DNMT3A (2; 3.4%), ASXL1 (2; 3.4%), SRSF2 (2; 3.4%), BCOR (1; 1.7%), TP53 (1; 1.7%) and ZRSR2 (1; 1.7%) (Figure 1A). JAK2 mutations were not detected in 89.0% (471/529) of our cohort. A diagnosis of PV was confirmed in 2 JAK2-negative patients based on clinical features and myeloid mutations were detected in both: SRSF2 and TET2 gene mutations in 1 patient and SRSF2, IDH2, ASXL1 gene mutations in the other patient. Three JAK2-negative patients tested positive for the BCR-ABL fusion and were diagnosed with chronic myeloid leukemia. The remaining 466 JAK2-negative patients were diagnosed with secondary erythrocytosis and myeloid mutations were found in 6% (28/466) of these cases. Mutations were detected in DNMT3A (12; 2.6%), TET2 (5; 1.1%), ASXL1 (5; 1.1%), TP53 (2; 0.4%), NF1 (2; 0.4%), KIT (1; 0.2%), U2AF1 (1; 0.2%) (Figure 1B). All patients with JAK2-negative secondary erythrocytosis had only one myeloid gene mutation detected.
Conclusion: Additional myeloid mutations other than JAK2 mutations are frequently identified in patients referred for erythrocytosis, with the highest frequencies observed in the TET2, DNMT3A and ASXL1 genes. The spectrum of myeloid mutations and overall incidence in JAK2-negative patients with secondary erythrocytosis is similar to the reported incidence of Clonal Hematopoiesis of Indeterminate Potential (CHIP) (Jaiswal et al., NEJM 2014), and suggests that these may represent incidental age-related mutations. By contrast, among the JAK2-positive patients, 34.5% had at least one additional myeloid mutation supporting a pathogenic role in these patients with myeloproliferative neoplasms. While concomitant myeloid mutations in patients with PV are well-described, further research is required to elucidate the significance of variants identified in JAK2-negative patients classified as secondary erythrocytosis in order to determine whether these mutations contribute to clinical phenotype or represent background CHIP.
No relevant conflicts of interest to declare.
Author notes
 This icon denotes a clinically relevant abstract
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal